Chapter 2 Field data
This chapter define sampled species and individuals. Specifically, this chapter details:
- Collected species
- Individuals from the METRADICA project
- Candidates with varying DBH and TWI
- Field maps
2.1 Species
We selected 10 species (Tab. 2.1) common with the METRADICA project representative of the tree phylogeny in Paracou (Fig. 2.1).
| Genus | Species |
|---|---|
| Casearia | javitensis |
| Chrysophyllum | prieurii |
| Conceveiba | guianensis |
| Gustavia | hexapetala |
| Jacaranda | copaia subsp. copaia |
| Laetia | procera |
| Protium | stevensonii |
| Tachigali | melinonii |
| Virola | michelii |
| Virola | surinamensis |
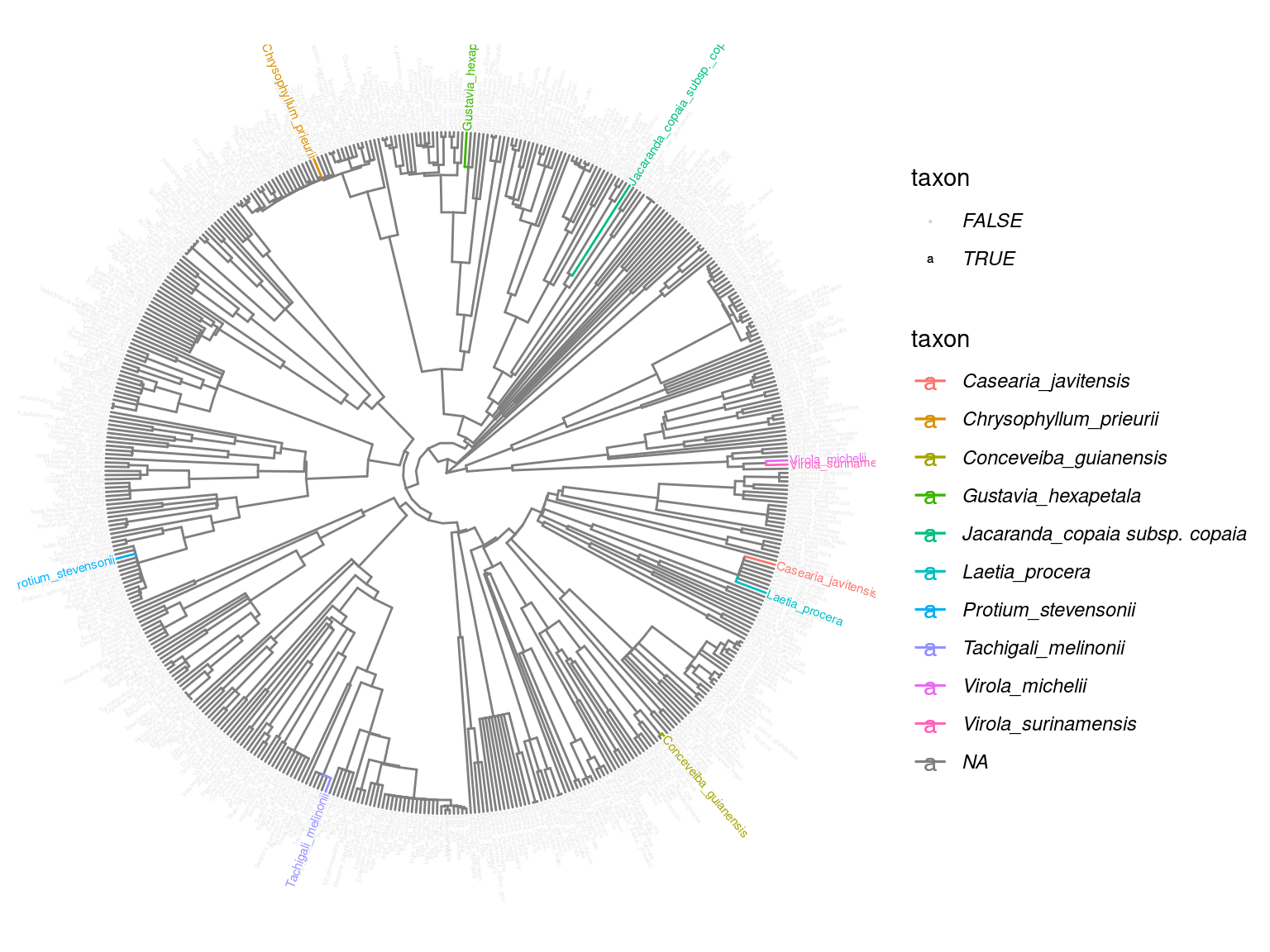
Figure 2.1: Selected species phylogeny.
2.2 METRADICA
This subparagraph details collected or wrong individuals in METRADICA (Tab. 2.2) and Marion’s candidates (Tab. 2.2, & Fig. 2.2), but candidates of the project will be focused on P16 independently from her candidates and adjusted in the field.
| TaxonActu | Candidate | Measured | Wrong |
|---|---|---|---|
| Casearia javitensis | 9 | 5 | 1 |
| Chrysophyllum prieurii | 56 | 12 | 5 |
| Conceveiba guianensis | 43 | 18 | 9 |
| Gustavia hexapetala | 87 | 4 | 4 |
| Jacaranda copaia subsp. copaia | 28 | 18 | 17 |
| Laetia procera | 10 | 9 | 1 |
| Protium stevensonii | 12 | 4 | NA |
| Tachigali melinonii | 34 | 18 | 4 |
| Virola michelii | 55 | 7 | NA |
| Virola surinamensis | 6 | 1 | NA |
| TaxonActu | 1 | 11 | 13 | 14 | 15 | 16 | 18(Guyaflux) | 6 |
|---|---|---|---|---|---|---|---|---|
| Casearia_javitensis | 1 | 3 | 4 | 1 | ||||
| Chrysophyllum_prieurii | 3 | 1 | 1 | 17 | 1 | |||
| Conceveiba_guianensis | 1 | 1 | 8 | 1 | ||||
| Gustavia_hexapetala | 23 | 5 | 9 | 4 | 5 | |||
| Jacaranda_copaia subsp. copaia | 3 | 2 | 3 | 4 | ||||
| Laetia_procera | 7 | 1 | 2 | |||||
| Protium_stevensonii | 1 | 1 | 6 | |||||
| Tachigali_melinonii | 2 | 1 | 8 | 1 | 2 | |||
| Virola_michelii | 1 | 1 | 23 | |||||
| Virola_surinamensis | 1 | 5 |
Figure 2.2: Marion’s candidates in P16.
2.3 Candidates
We selected 15 individuals per species in P16 (10 + 5 extras) minimizing DBH variation while maximizing TWI variation (Fig. 2.3, Tab. 2.4, Fig. 2.4). We are lacking individuals for Casearia javitensis, Protium stevensonii, and Virola surinamensis.
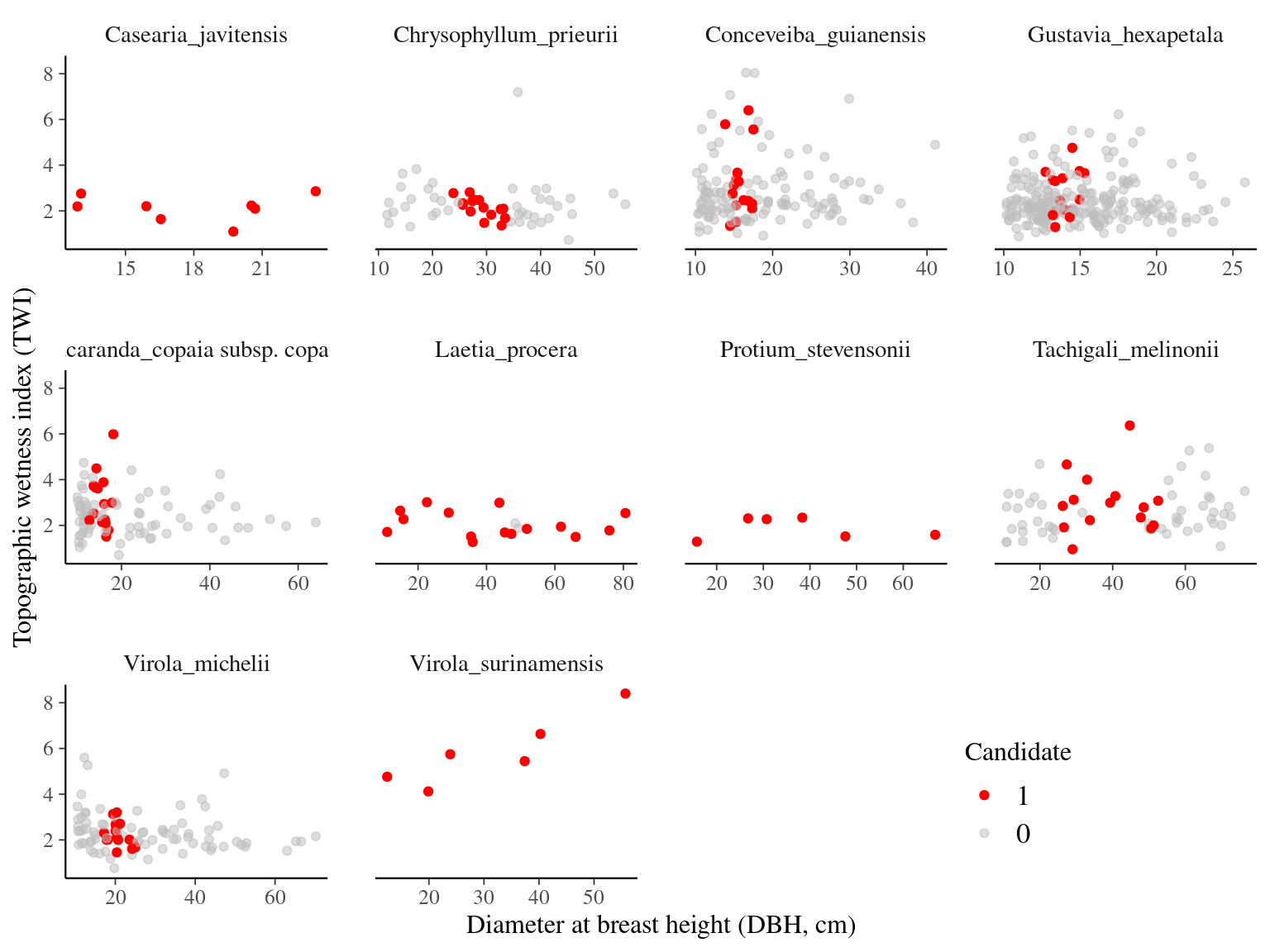
Figure 2.3: DBH and TWI of candidates in P16.
| taxon | Nsel | N |
|---|---|---|
| Casearia_javitensis | 8 | 8 |
| Chrysophyllum_prieurii | 15 | 55 |
| Conceveiba_guianensis | 15 | 133 |
| Gustavia_hexapetala | 15 | 238 |
| Jacaranda_copaia subsp. copaia | 15 | 88 |
| Laetia_procera | 15 | 17 |
| Protium_stevensonii | 6 | 6 |
| Tachigali_melinonii | 15 | 64 |
| Virola_michelii | 15 | 92 |
| Virola_surinamensis | 6 | 6 |
Figure 2.4: Candidates in P16.
2.4 Maps
Maps were automatically built using sf and ggplot2 and save in the folder maps:
contour <- st_read("data/ContourLinesPlots/ContourLinePlots.shp", quiet = T)
crs_rot = "+proj=omerc +lat_0=36.934 +lonc=-90.849 +alpha=0 +k_0=.7 +datum=WGS84 +units=m +no_defs +gamma=20"
make_map <- function(file, title, subplots){
sublimits <- filter(limits, Plot == 16, Subplot %in% subplots) %>%
st_transform(crs = crs_rot)
subcontour <- st_transform(contour, crs = crs_rot) %>% st_crop(sublimits)
subcandidates <- filter(candidates, candidate == 1, SubPlot %in% subplots) %>%
st_transform(crs = crs_rot) %>%
mutate(label = paste0("P", Plot, "-", SubPlot, "-", TreeFieldNum, "_", substr(Genus, 1, 3), substr(Species, 1, 3), "_", round(DBH)))
g <- ggplot() +
geom_sf(data = subcontour, fill = NA, col = "lightgrey") +
geom_sf(data = sublimits, fill = NA, col = "darkgrey", aes(text = subplots)) +
geom_sf_text(data = sublimits, aes(label = Subplot), colour = "darkgrey") +
geom_sf(data = subcandidates, col = "black") +
ggrepel::geom_text_repel(
data = subcandidates,
aes(label = label, geometry = geometry),
stat = "sf_coordinates",
min.segment.length = 0
) +
theme(axis.title = element_blank(), axis.text = element_blank(),
axis.ticks = element_blank(), axis.line = element_blank()) +
scale_color_discrete(guide = "none") +
ggtitle(title)
ggsave(g, file = file, path = 'maps', width = 297, height = 420, unit = 'mm', dpi = 300)
}2.5 Extra FTH
Candidate individuals from the P16 were not enough for the FTH field campaign. We added individuals from the P6:
2.6 Extra Post - FTH
Individuals were still missing after the FTH field campaign. We added individuals from the P16:
2.7 Leaf variation
For the leaf variation within individual we sampled across the 27 Virola michelii from the P16 that followed our requirements (DBH between 30th and 60th quantiles):
Figure 2.5: Candidates in P16.