Chapter 14 Manuscript figures
This chapter develop the code for figure preparation for the final manuscript.
14.0.1 Figure 1

Figure 14.1: Fig. 1 | Crown mutations and transmitted mutations in the genomic landscape of Angela and Sixto assembled pseudo-chromosomes. The genomic landscape is similarly portrayed for the two tropical trees: the Dicorynia guianensis tree named Angela (a), and the Sextonia rubra tree named Sixto (b). The first (most external) track represents the percentage of GC in the whole genome with the black line and in the transposable elements with the green line. The second (least external) track represents the percentage of transposable elements with purple bars. The third track (middle) represents the percentage of genes with blue bars. The fourth (least internal) track represents the number of somatic mutations detected in the tree crown with yellow bars. The fifth (innermost) track represents the allelic fraction of the somatic mutations detected in the tree crown in yellow and the somatic mutations transmitted to the embryos in red. The inner labels indicate the type of mutations for somatic mutations transmitted to embryos. All measurements are calculated in non-overlapping windows of 100 kb. A ruler is drawn on each pseudo-chromosome, with tick marks every 2 Mb.
14.0.2 Figure 2
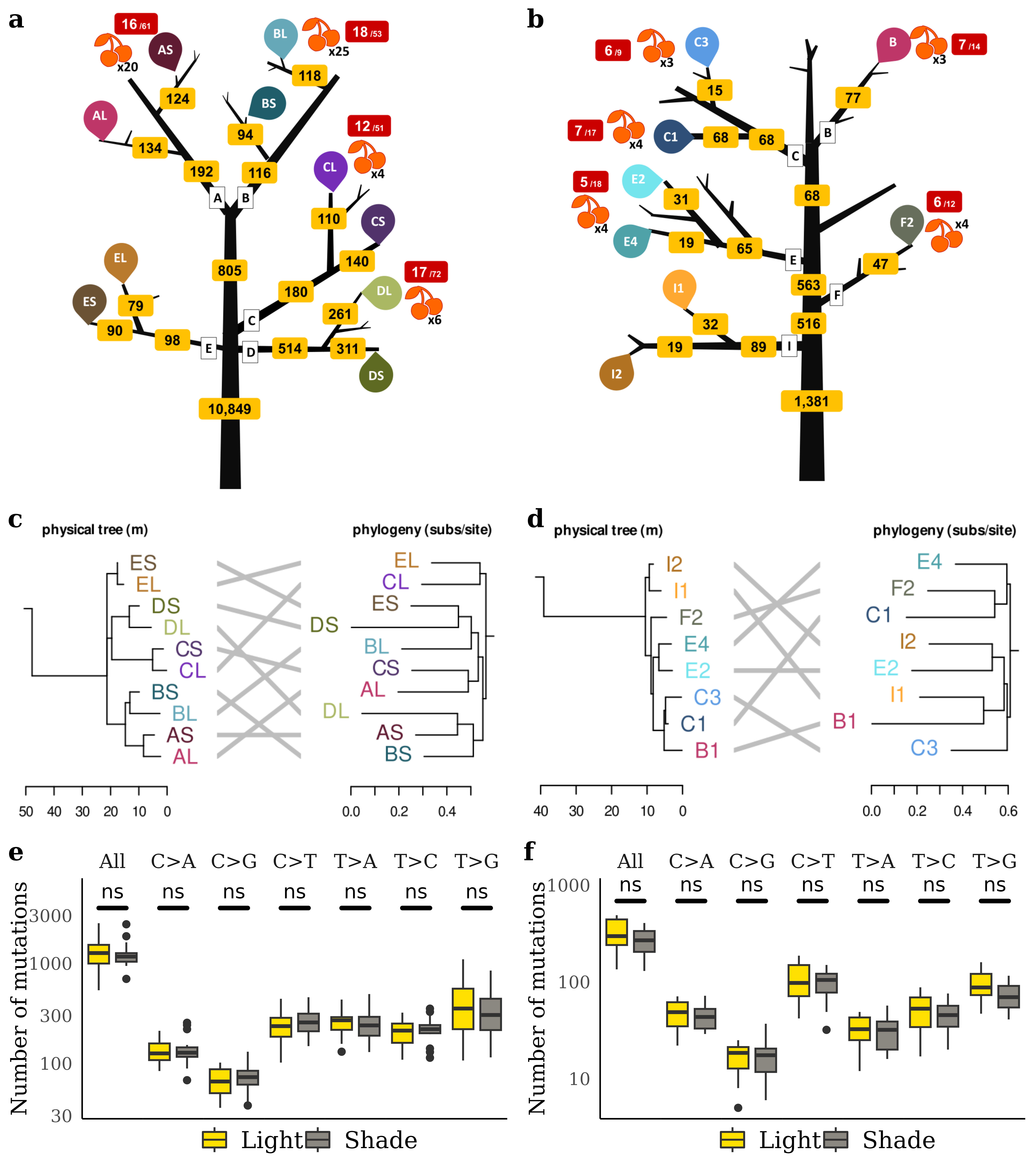
Figure 14.2: Fig. 2 | Somatic mutation distributions through physical trees, phylogenies, and with light. Somatic mutation distributions through physical trees, phylogenies, and with light are similarly portrayed for the two tropical trees: the Dicorynia guianensis tree named Angela (a,c,e), and the Sextonia rubra tree named Sixto (b,d,f). (a-b) The physical architecture of the tree is shown in black with the bough names in white boxes. The number of somatic mutations through the crown is indicated in the yellow boxes before the original branching event. The balloons circles indicate the sample points of three leaves in the light-exposed branches (light colours) and in the shaded branches (dark colours). Fruit sampling points are represented by red fruits, with the number of fruits sampled indicated in black. The red boxes with white labels indicate the transmission of mutations to fruit embryos out of the total number of mutations tested. (c-d) A side-by-side comparison of the physical tree (left, branch length in metres) and the maximum likelihood phylogeny (right, branch length in substitutions per site). The letters on the ends of the branches indicate the sample points with unique colours. (e-f) The effect of light exposure on the accumulation of somatic mutations as a function of mutation type is represented in yellow and grey boxes. The yellow boxes represent the number of mutations accumulated in all leaves of light exposed branches and the grey boxes in all leaves of shaded branches. The ‘ns’ labels indicate non-significant differences in Student’s t-tests. Mutation types include all mutations and all types of transitions and transversions. The y-axis has been scaled logarithmically to facilitate reading of low values.
14.1 Figure 3
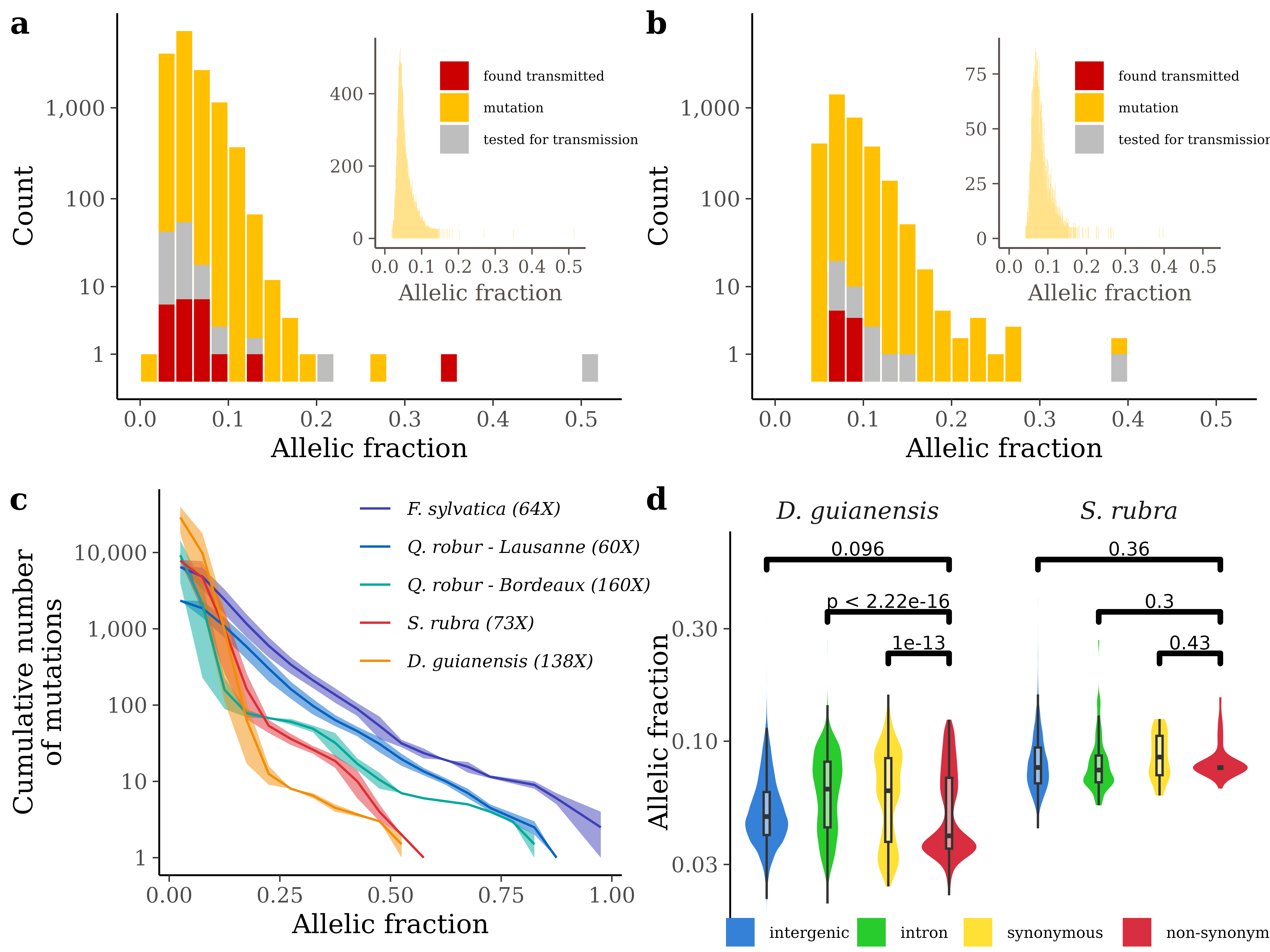
Figure 14.3: Fig. 3 | Allelic fractions of somatic mutations among trees and among genomic elements. Histogram of allelic fractions of mutations detected in the crown of the two tropical trees: the Dicorynia guianensis tree named Angela (a), and the Sextonia rubra tree named Sixto (b). The main histogram shows the allelic fractions of the somatic mutations using a bin of 0.02 and a log-transformed count with the mutations detected in the crown in yellow, the mutations tested for transmission in grey, and the mutations transmitted to the embryos in red. The inner histogram shows the allelic fractions of the somatic mutations using a bin of 0.001 and a natural count. (c) Cumulative number of somatic mutations per branch with decreasing allelic fraction for five trees reanalysed with the same pipeline. The five trees include the two tropical trees studied, the Dicorynia guianensis tree named Angela in orange and the Sextonia rubra tree named Sixto in red, and three temperate trees, two pedunculate oaks Quercus robur named 3P in green and Napoleon in blue and a tortuous beech Fagus sylvatica named Verzy in purple. All trees were analysed with the same pipeline (see methods) but were sequenced with a different depth indicated in brackets. The line represents the median value while the area represents the minimum and maximum values on the 2 to 10 branches per tree. (d) Comparisons of allelic fractions for non-synonymous mutations in red with synonymous mutations in yellow, intronic mutations in green and intergenic mutations in blue for the two tropical trees: the Dicorynia guianensis tree named Angela (left panel), and the Sextonia rubra tree named Sixto (right panel). The p-value above the bars indicates the significance of the Student’s T-test for the pairs of groups.