Calibration of forest structure parameters (crown radius allometry and mortality) in TROLL 4.0.1 (as mortality differs between TROLL 3.1.8 and 4.0.1):
\(m\) : minimal death rate in death per year calibrated with TROLL 3.1.8 to 0.055 both at Paracou and Tapajos\(a_{CR}\) : crown radius intercept calibrated with TROLL 3.1.8 to 1.85 at Paracou and to 2.50 at Tapajos\(b_{CR}\) : crown radius slope calibrated with TROLL 3.1.8 to 0.4445 at Paracou and to 0.8150 at Tapajos
Parameters spaces
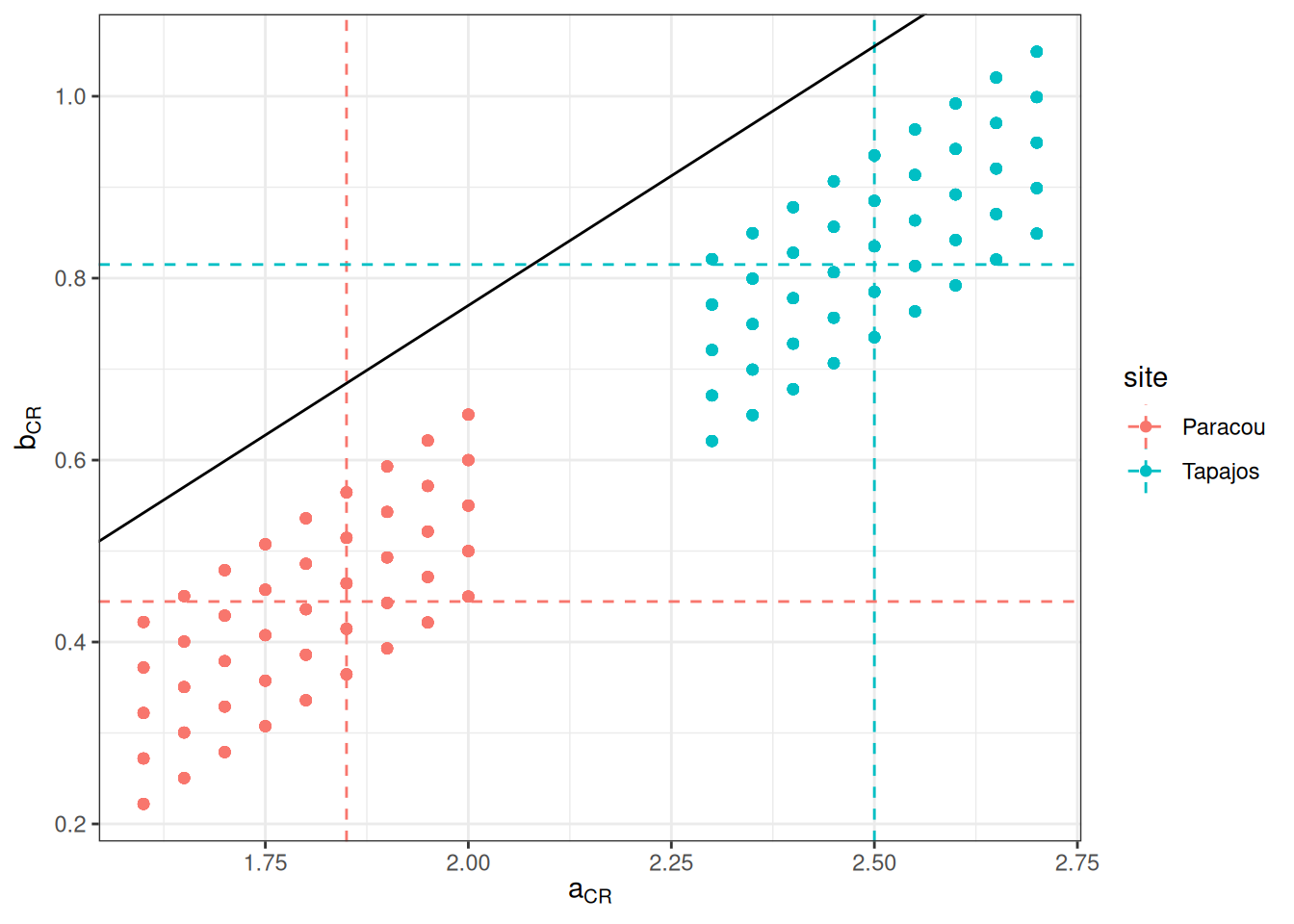
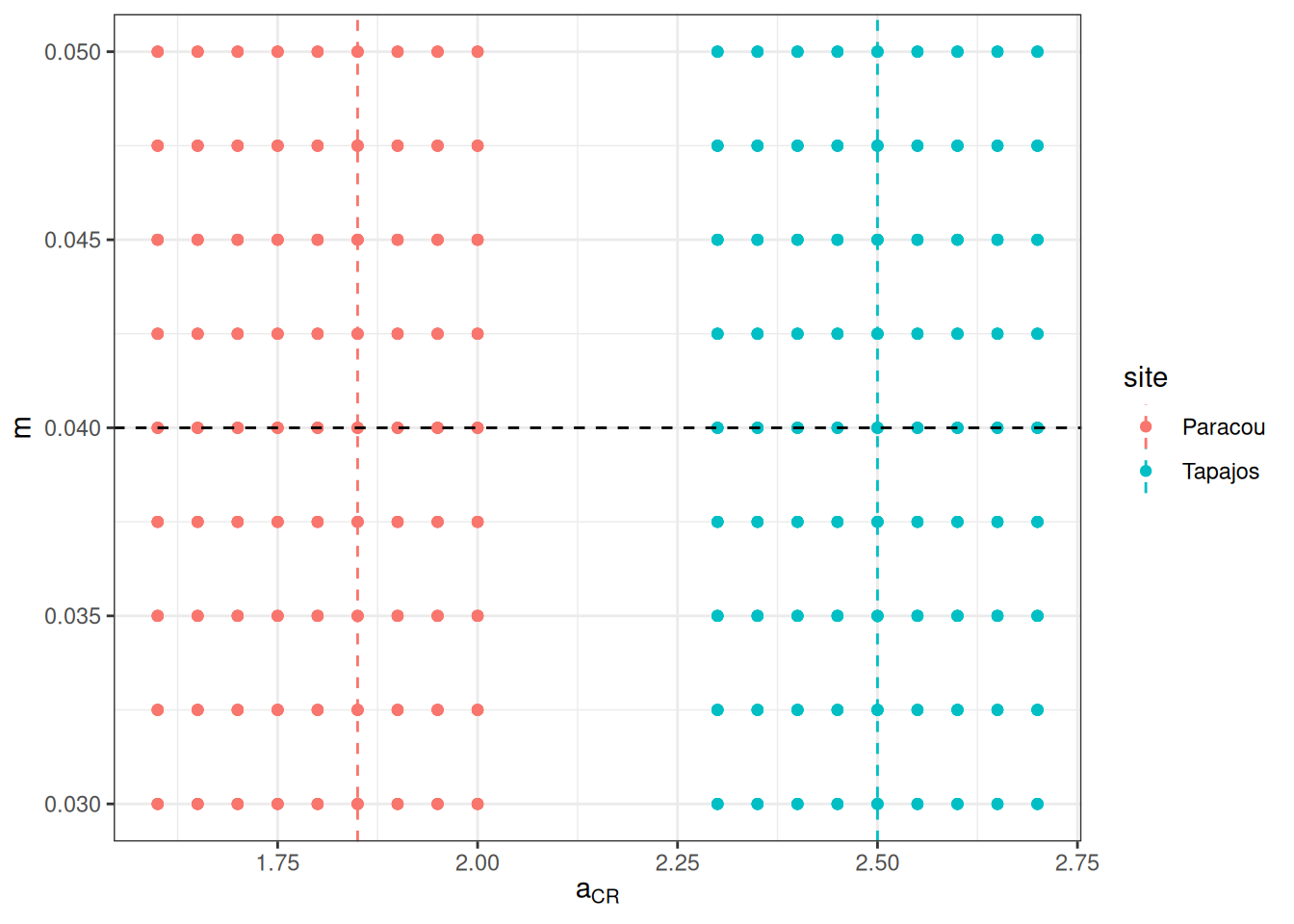
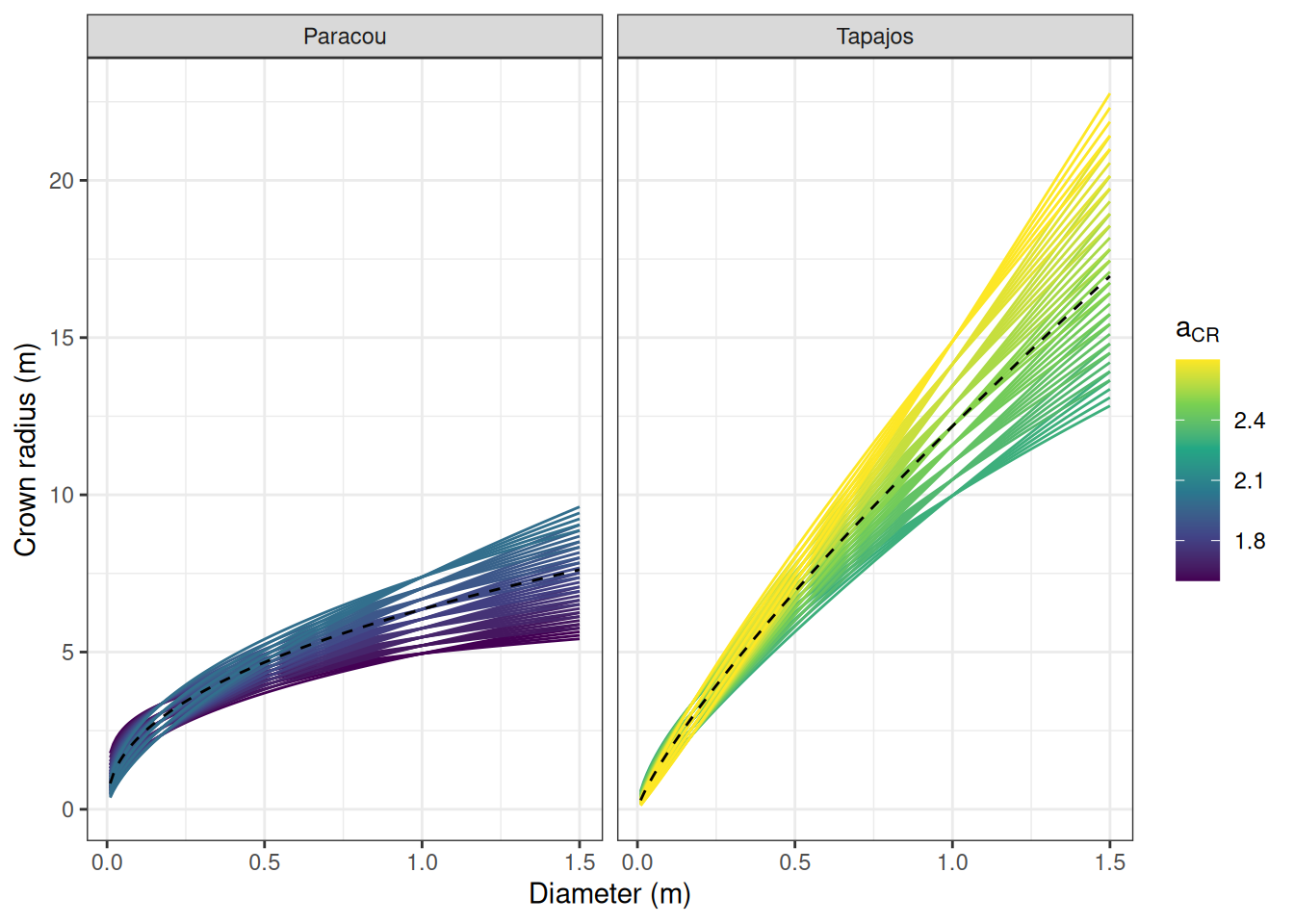
We prepared the parameter space for calibration defining low limit, high limit, and step for a, m, and the error of b for Paracou and Tapajos independently using calibration of forest crown radius allometry from TROLL 3.1.8 as prior and tests with TROLL 4.0.1 for the mortality prior. We obtained a calibration grid of 810 simulations . Dashed lines shows prior values. The last figure shows the corresponding projected allometries of crown radius.
Code
<- data.frame (site = "Paracou" ,parameter = c ("a" , "error_b" , "m" ),low = c (1.6 , - 6 * 0.05 , 0.03 ),high = c (2.0 , - 2 * 0.05 , 0.05 ),by = c (0.05 , 0.05 , 0.0025 ))<- data.frame (site = "Tapajos" ,parameter = c ("a" , "error_b" , "m" ),low = c (2.3 , - 6 * 0.05 , 0.03 ),high = c (2.7 , - 2 * 0.05 , 0.05 ),by = c (0.05 , 0.05 , 0.0025 ))<- bind_rows (pars_par, pars_tap)<- data.frame (a = seq (pars_par$ low[1 ], pars_par$ high[1 ], by = pars_par$ by[1 ])) %>% mutate (error_b = list (seq (pars_par$ low[2 ], pars_par$ high[2 ], by = pars_par$ by[2 ]))) %>% unnest (error_b) %>% mutate (b = - 0.39 + 0.57 * a + error_b) %>% mutate (m = list (seq (pars_par$ low[3 ], pars_par$ high[3 ], by = pars_par$ by[3 ]))) %>% unnest (m) %>% mutate (site = "Paracou" )<- data.frame (a = seq (pars_tap$ low[1 ], pars_tap$ high[1 ], by = pars_tap$ by[1 ])) %>% mutate (error_b = list (seq (pars_tap$ low[2 ], pars_tap$ high[2 ], by = pars_tap$ by[2 ]))) %>% unnest (error_b) %>% mutate (b = - 0.39 + 0.57 * a + error_b) %>% mutate (m = list (seq (pars_tap$ low[3 ], pars_tap$ high[3 ], by = pars_tap$ by[3 ]))) %>% unnest (m) %>% mutate (site = "Tapajos" )<- bind_rows (grid_par, grid_tap)
Code
Paracou
a
1.60
2.00
0.0500
Paracou
error_b
-0.30
-0.10
0.0500
Paracou
m
0.03
0.05
0.0025
Tapajos
a
2.30
2.70
0.0500
Tapajos
error_b
-0.30
-0.10
0.0500
Tapajos
m
0.03
0.05
0.0025
Code
%>% ggplot (aes (a, b, col = site)) + geom_point () + geom_vline (aes (xintercept = a, col = site), linetype = "dashed" , data = data.frame (a = c (1.85 , 2.50 ), site = c ("Paracou" , "Tapajos" ))) + geom_hline (aes (yintercept = b, col = site), linetype = "dashed" , data = data.frame (b = c (0.4445 , 0.8150 ), site = c ("Paracou" , "Tapajos" ))) + geom_abline (intercept = - 0.37 , slope = 0.57 , col = "black" ) + theme_bw () + xlab (expression (a[CR])) + ylab (expression (b[CR]))
Code
%>% ggplot (aes (a, m, col = site)) + geom_point () + geom_vline (aes (xintercept = a, col = site), linetype = "dashed" , data = data.frame (a = c (1.85 , 2.50 ), site = c ("Paracou" , "Tapajos" ))) + geom_hline (yintercept = 0.040 , linetype = "dashed" ) + theme_bw () + xlab (expression (a[CR])) + ylab (expression (m))
Code
%>% mutate (dbh = list (seq (1 , 150 , 1 )/ 100 )) %>% unnest (dbh) %>% mutate (cr = exp (a+ b* log (dbh))) %>% ggplot (aes (dbh, cr)) + geom_line (aes (col = a, group = paste (a, b))) + theme_bw () + scale_colour_viridis_c (expression (a[CR])) + xlab ("Diameter (m)" ) + ylab ("Crown radius (m)" ) + geom_line (col = "black" , linetype = "dashed" ,data = bind_rows (data.frame (site = "Paracou" , dbh = seq (1 , 150 , 1 )/ 100 ) %>% mutate (cr = exp (1.85+0.4445 * log (dbh))),data.frame (site = "Tapajos" , dbh = seq (1 , 150 , 1 )/ 100 ) %>% mutate (cr = exp (2.50+0.8150 * log (dbh)))+ facet_wrap (~ site)
Observations
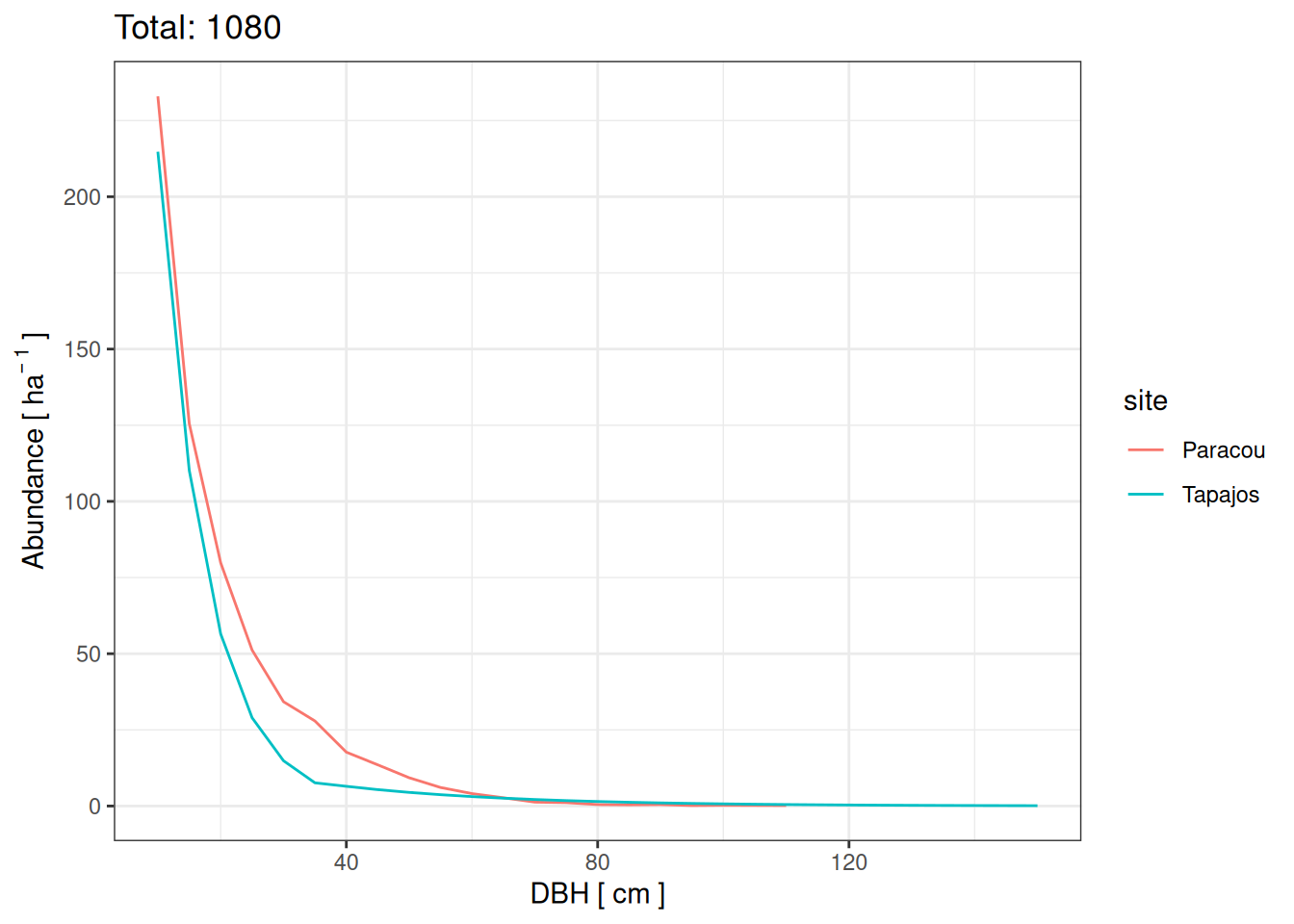
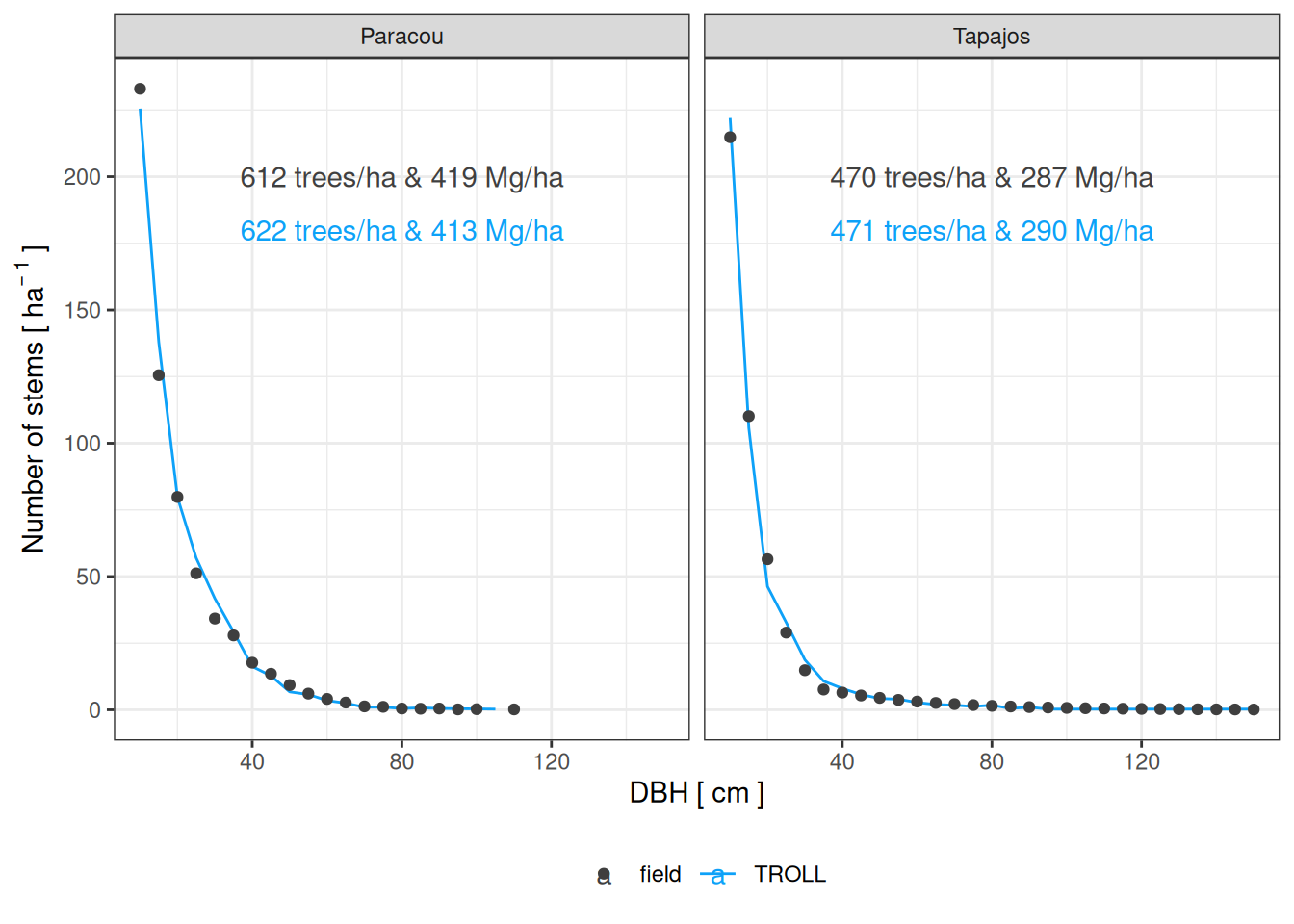
We used different evaluation metrics per site to rely on a single data source and minimise discrepancies.
For Tapajos and according to Rice et al. (2004 ) :
470 live trees per hectare with diameter at breast height (dbh) ≥10
Aboveground live biomass was 143.7 ± 5.4 Mg C/ha
Stem density -0.058 (±0.003) dbh_cm + 2.912 (±0.076) for dbh < 40
Stem density -0.016 (±0.001) dbh_cm + 1.451 (±0.04) for dbh > 40
Stem density is shown below
For Paracou according to the inventories:
612 ± 37 live trees per hectare with diameter at breast height (dbh) ≥10 (from inventory)
Basal area was 30.8 ± 0.836 m2/ha
Stem density is shown below
Code
<- read_tsv ("outputs/inventories.tsv" , show_col_types = FALSE ) %>% filter (site == "Paracou" , type == "default" ) %>% filter (dbh > 10 ) %>% group_by (plot) %>% summarise (abundance = n ()/ unique (plot_size)) %>% ungroup ()# paste(round(median(abd_par$abundance)), "±", round(sd(abd_par$abundance))) # read_tsv("outputs/inventories.tsv") %>% # filter(site == "Paracou", type == "default", dbh >= 10) %>% # group_by(plot) %>% # summarise(ba = sum((dbh/2)^2*pi)/10^4/unique(plot_size)) %>% # summarise(ba_mean = median(ba), ba_sd = sd(ba)) <- data.frame (dbh_class = seq (10 , 150 , 5 )) %>% mutate (abundance = ifelse (dbh_class < 40 , 10 ^ (- 0.058 * dbh_class+2.912 ), 10 ^ (- 0.016 * dbh_class+1.451 ))) %>% mutate (site = "Tapajos" )<- read_tsv ("outputs/inventories.tsv" , show_col_types = FALSE ) %>% filter (site == "Paracou" , type == "default" ) %>% filter (dbh > 10 ) %>% mutate (dbh_class = cut (dbh, breaks = c (seq (10 , 155 , by = 5 )), labels = c (seq (10 , 150 , by = 5 )))) %>% mutate (dbh_class = as.numeric (as.character (dbh_class))) %>% group_by (plot, dbh_class) %>% summarise (abundance = n ()/ unique (plot_size)) %>% group_by (dbh_class) %>% summarise (abundance = median (abundance)) %>% mutate (site = "Paracou" )# obs <- bind_rows(obs_tap, obs_par) <- bind_rows (obs_tap, obs_par)ggplot (obs, (aes (dbh_class, abundance, col = site))) + geom_line () + theme_bw () + xlab ("DBH [ cm ]" ) + ylab (expression (Abundance~ "[" ~ ha^ {- ~ 1 }~ "]" )) + ggtitle (paste ("Total:" , round (sum (obs$ abundance))))
Parameters effect
Code
bind_rows (read_tsv ("results/calib_str4_Paracou.tsv" ),read_tsv ("results/calib_str4_Tapajos.tsv" )%>% mutate (dbh_class = cut (dbh_cm, breaks = seq (10 , 205 , by = 5 ), labels = seq (10 , 200 , by = 5 ),include.lowest = TRUE )) %>% mutate (dbh_class = as.numeric (as.character (dbh_class))) %>% group_by (site, a, b, m, dbh_class) %>% summarise (abundance = n ()/ 4 , agb = sum (agb)/ 4 , ba = sum ((dbh_cm/ 2 )^ 2 * pi)/ 10 ^ 4 / 4 ) %>% write_tsv ("outputs/calib_str4.tsv" )
\(a_{CR}\)
Code
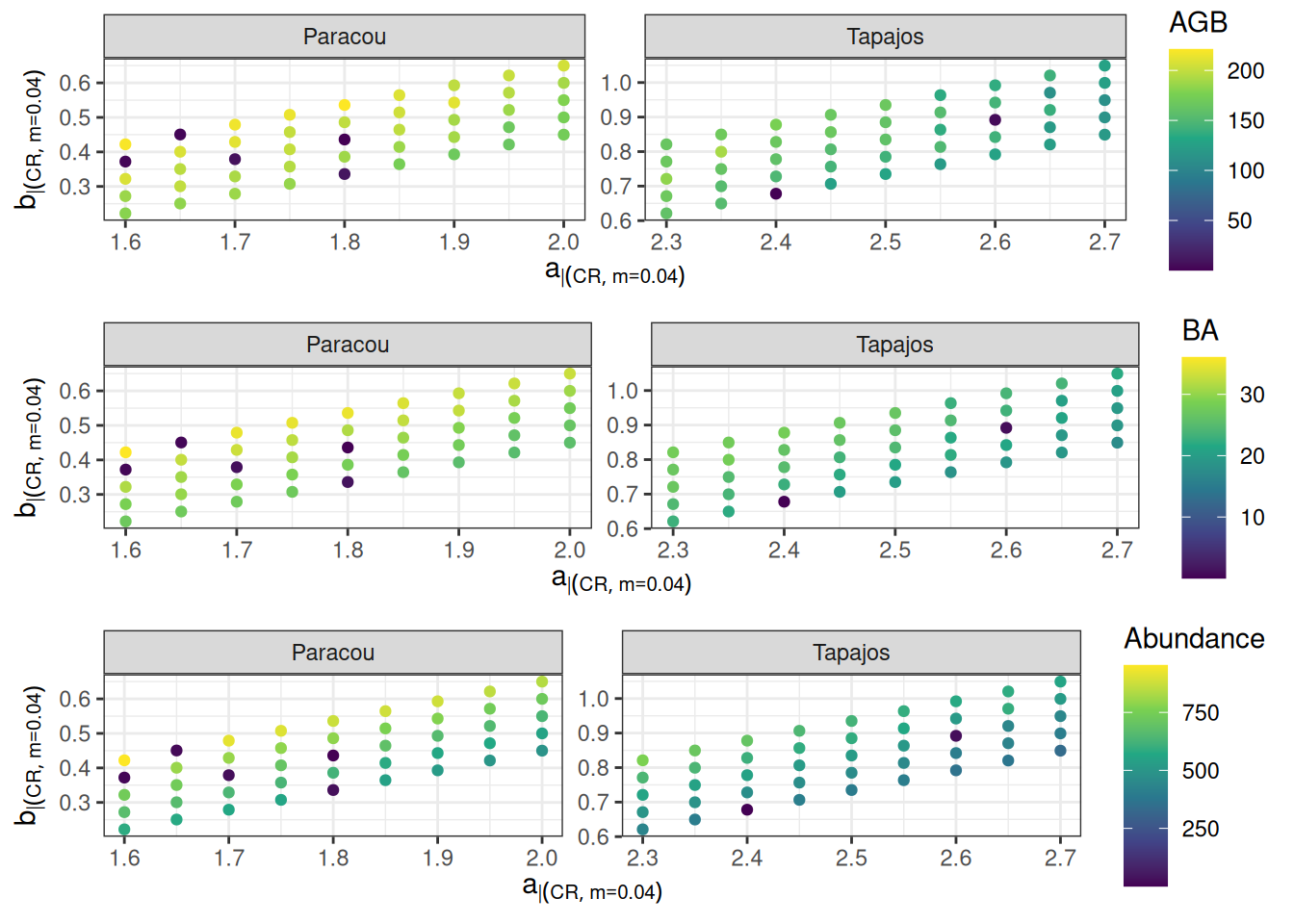
<- read_tsv ("outputs/calib_str4.tsv" ) %>% group_by (site, a, b, m) %>% summarise (agb = sum (agb)/ 10 ^ 3 / 2 ,ba = sum (ba),abundance = sum (abundance)) %>% filter (m == 0.040 ):: plot_grid (%>% ggplot (aes (a, b, col = agb)) + geom_point () + facet_wrap (~ site, scales = "free" ) + theme_bw () + scale_color_viridis_c ("AGB" ) + xlab (expression (a[CR| m== 0.04 ])) + ylab (expression (b[CR| m== 0.04 ])),%>% ggplot (aes (a, b, col = ba)) + geom_point () + facet_wrap (~ site, scales = "free" ) + theme_bw () + scale_color_viridis_c ("BA" ) + xlab (expression (a[CR| m== 0.04 ])) + ylab (expression (b[CR| m== 0.04 ])),%>% ggplot (aes (a, b, col = abundance)) + geom_point () + facet_wrap (~ site, scales = "free" ) + theme_bw () + scale_color_viridis_c ("Abundance" ) + xlab (expression (a[CR| m== 0.04 ])) + ylab (expression (b[CR| m== 0.04 ])), nrow = 3 )
Code
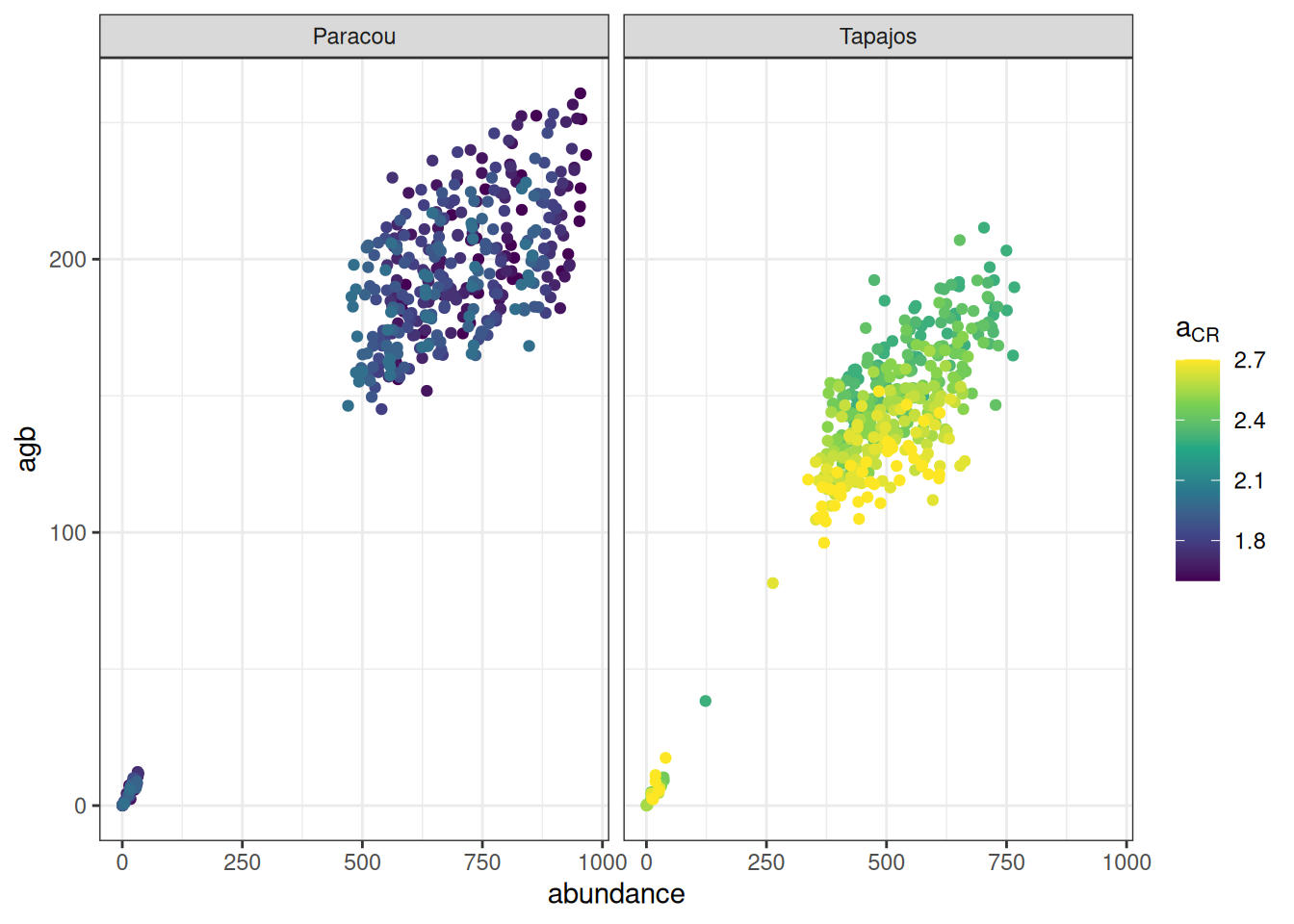
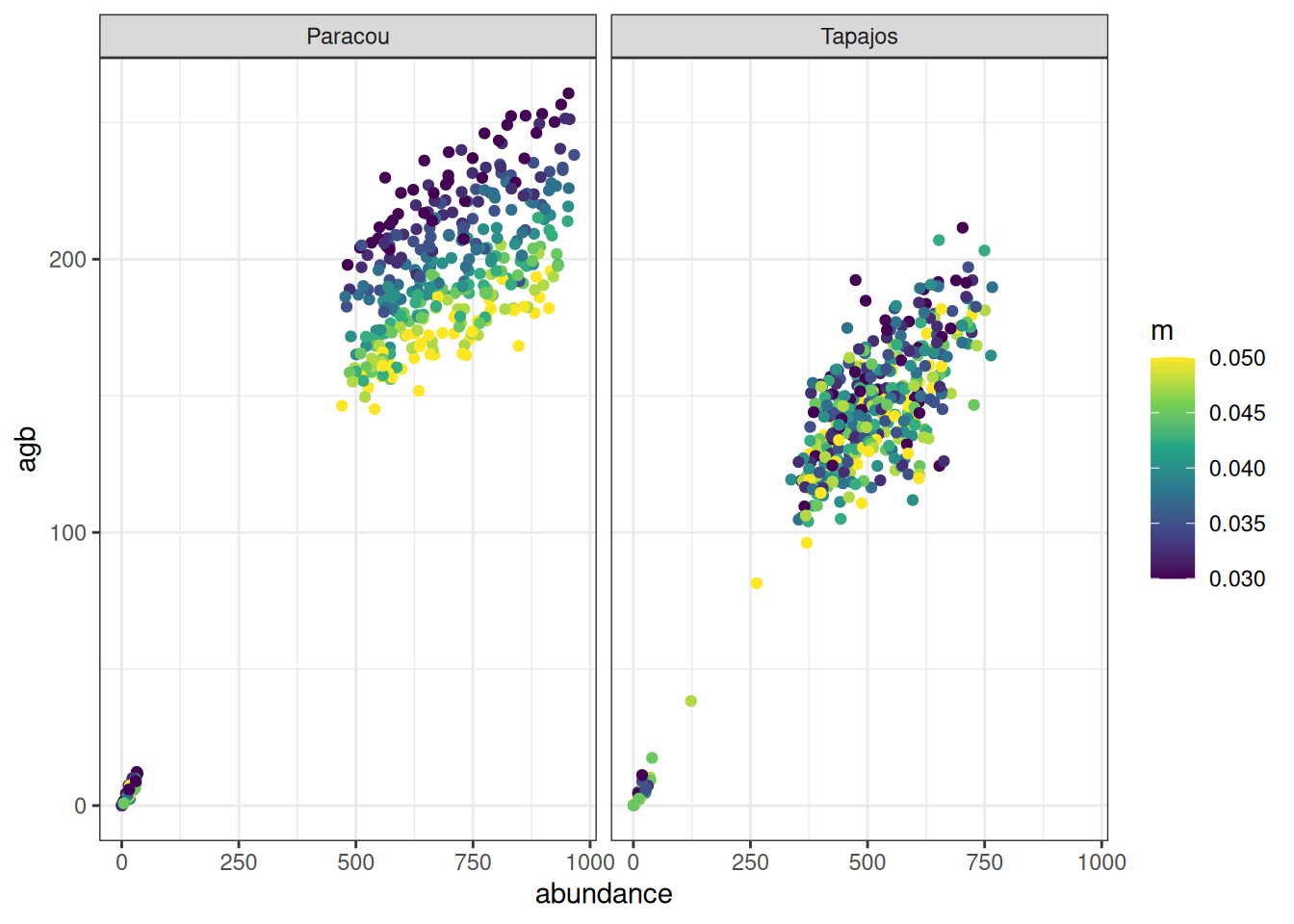
read_tsv ("outputs/calib_str4.tsv" ) %>% group_by (site, a, b, m) %>% summarise (abundance = sum (abundance), agb = sum (agb)/ 10 ^ 3 / 2 ) %>% ggplot (aes (abundance, agb, col = a)) + geom_point () + theme_bw () + scale_color_viridis_c (expression (a[CR])) + facet_wrap (~ site)
Code
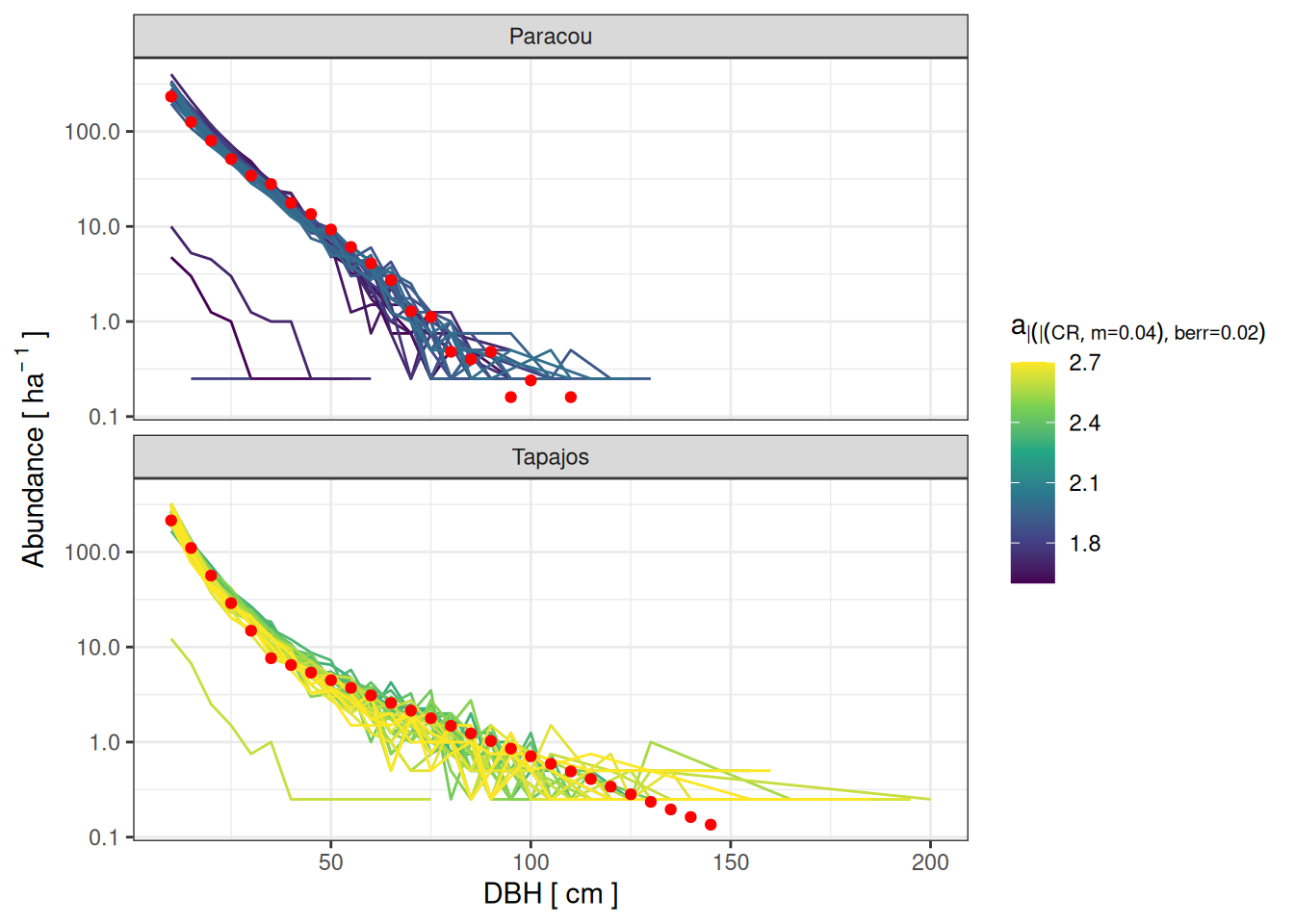
read_tsv ("outputs/calib_str4.tsv" ) %>% filter (m == 0.040 ) %>% mutate (berr = - 0.39 + 0.57 * a - b) %>% filter (berr > 0.1 , berr < 0.3 ) %>% ggplot (aes (dbh_class, abundance)) + geom_line (aes (col = a, group = paste (a, b, m))) + geom_point (data = filter (obs, dbh_class < 150 ), col = "red" ) + scale_y_log10 (labels = scales:: comma) + xlab ("DBH [ cm ]" ) + ylab (expression (Abundance~ "[" ~ ha^ {- ~ 1 }~ "]" )) + scale_color_viridis_c (expression (a[CR| m== 0.04 | berr== 0.02 ])) + theme_bw () + facet_wrap (~ site, nrow = 2 )
\(b_{CR}\)
Code
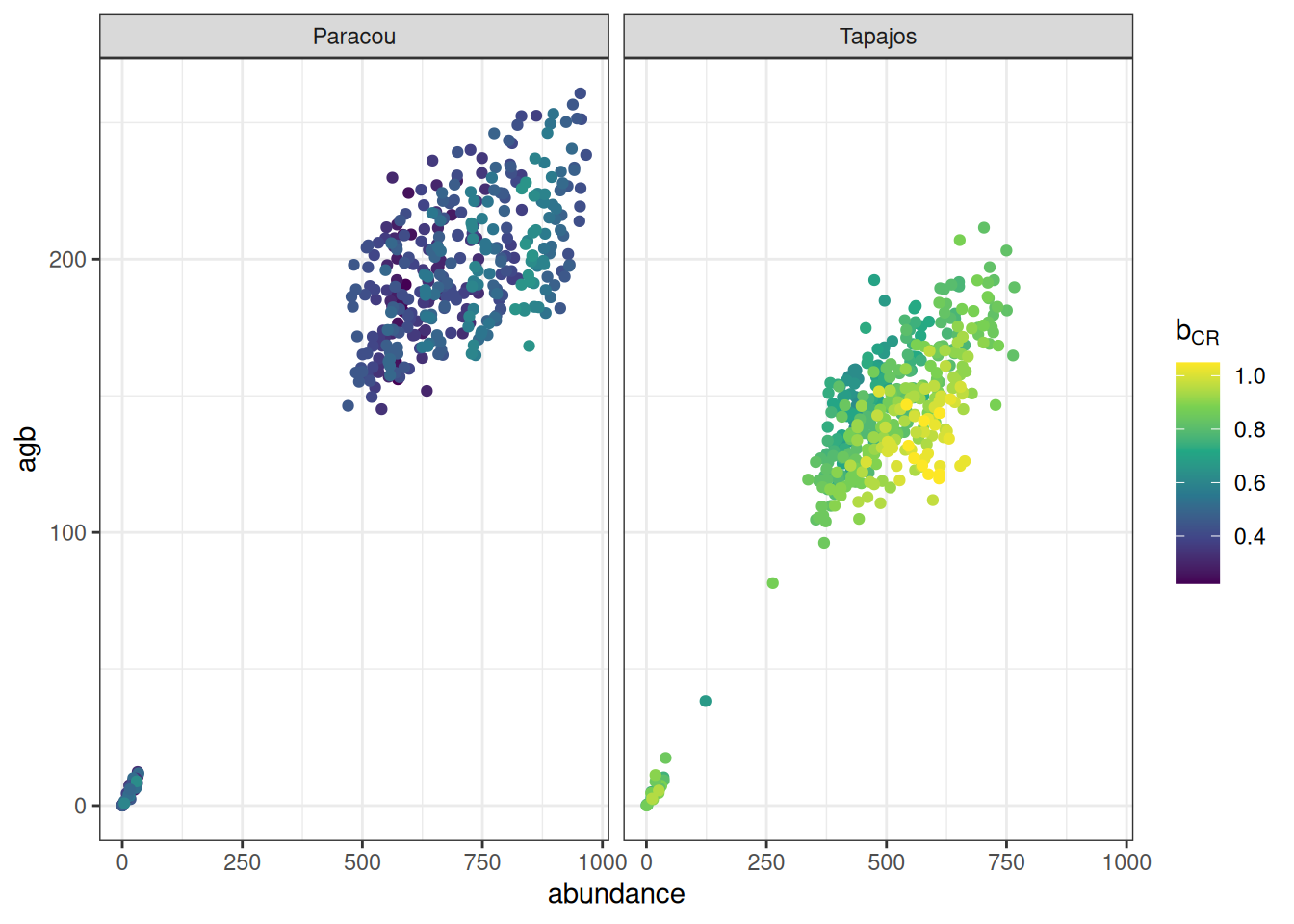
read_tsv ("outputs/calib_str4.tsv" ) %>% group_by (site, a, b, m) %>% summarise (abundance = sum (abundance), agb = sum (agb)/ 10 ^ 3 / 2 ) %>% ggplot (aes (abundance, agb, col = b)) + geom_point () + theme_bw () + scale_color_viridis_c (expression (b[CR])) + facet_wrap (~ site)
\(m\)
Code
read_tsv ("outputs/calib_str4.tsv" ) %>% group_by (site, a, b, m) %>% summarise (abundance = sum (abundance), agb = sum (agb)/ 10 ^ 3 / 2 ) %>% ggplot (aes (abundance, agb, col = m)) + geom_point () + theme_bw () + scale_color_viridis_c (expression (m)) + facet_wrap (~ site)
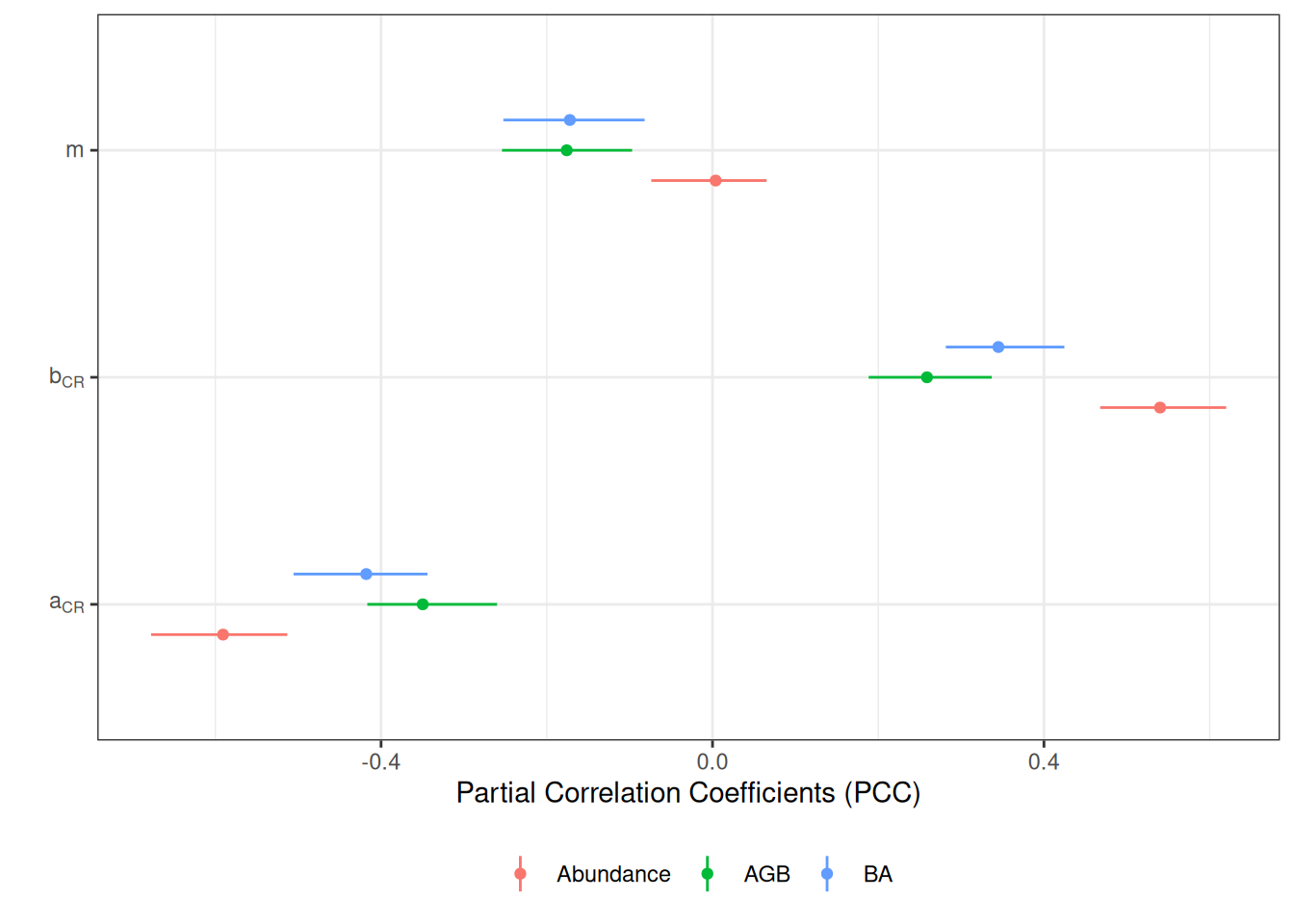
Sensitivity to parameters
Code
<- read_tsv ("outputs/calib_str4.tsv" ) %>% group_by (site, a, b, m) %>% summarise (abundance = sum (abundance), agb = sum (agb), ba = sum (ba))<- lapply (c ("abundance" , "agb" , "ba" ), function (var) :: pcc (t[c ("a" , "b" , "m" )], t[var], nboot = 100 )$ PCC %>% rownames_to_column (var = "parameter" ) %>% mutate (variable = var)) %>% bind_rows ()
Code
<- sens %>% mutate (parameter = recode (parameter,"a" = "a[CR]" ,"b" = "b[CR]" ,"m" = "m" )) %>% mutate (variable = recode (variable,"abundance" = "Abundance" ,"agb" = "AGB" ,"ba" = "BA" )) %>% ggplot (aes (parameter, original, col = variable)) + geom_point (position = position_dodge (0.4 )) + theme_bw () + coord_flip () + geom_linerange (aes (ymin = ` min. c.i. ` , ymax = ` max. c.i. ` ), position = position_dodge (0.4 )) + ylab ("Partial Correlation Coefficients (PCC)" ) + xlab ("" ) + theme (legend.position = "bottom" ) + scale_x_discrete (labels = scales:: label_parse ()) + scale_colour_discrete ("" )
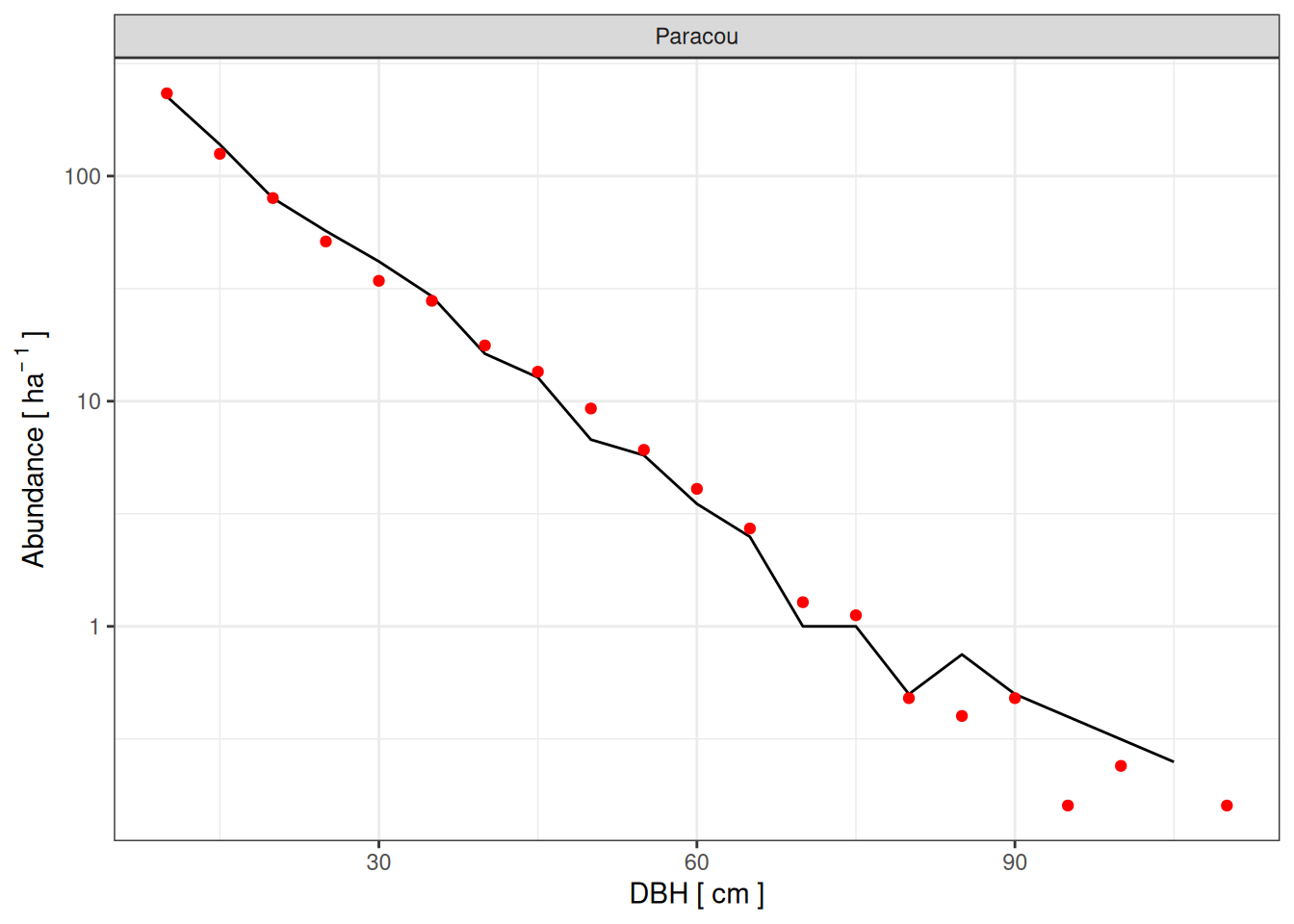
Best parameters
Paracou
Code
<- read_tsv ("outputs/calib_str4.tsv" ) %>% filter (site == "Paracou" ) %>% select (site, a, b, m) %>% unique () %>% mutate (dbh_class = list (seq (10 , 150 , by = 5 ))) %>% unnest (dbh_class) %>% left_join (read_tsv ("outputs/calib_str4.tsv" )) %>% select (- agb) %>% mutate_at (c ("abundance" , "ba" ), ~ ifelse (is.na (.), 0 , .)) %>% rename (simulated = abundance) %>% left_join (rename (obs, observed = abundance)) %>% mutate (observed = ifelse (is.na (observed), 0 , observed)) %>% group_by (site, a, b, m) %>% summarise (rmsep_dist = sqrt (mean ((observed - simulated)^ 2 )),rmsep_ba = sqrt ((30.8 - sum (ba))^ 2 ),rmsep_abund = sqrt ((612 - sum (simulated))^ 2 )%>% ungroup () %>% mutate (all_rmsep_norm = rmsep_dist/ 30.5 + rmsep_ba/ 30.8 + rmsep_abund/ 612 ) %>% mutate (sim = paste0 (site, "_" , a, "_" , b, "_" , m))<- eval_par %>% group_by (site) %>% slice_min (all_rmsep_norm, n = 5 )
Code
read_tsv ("outputs/calib_str4.tsv" ) %>% mutate (sim = paste0 (site, "_" , a, "_" , b, "_" , m)) %>% left_join (best_par) %>% filter (! is.na (all_rmsep_norm)) %>% group_by (site, a, b, m) %>% summarise (abundance = sum (abundance), ba = sum (ba)) %>% gather (variable, value, - site) %>% group_by (site, variable) %>% summarise (median = median (value), minimum = min (value), maximum = max (value)) %>% :: kable (caption = "Best structure parameters according to calibration with inventory data." )
Best structure parameters according to calibration with inventory data.
Paracou
a
1.8000
1.80000
1.90000
Paracou
abundance
606.2500
587.50000
628.00000
Paracou
b
0.3860
0.38600
0.44300
Paracou
ba
30.0482
29.46175
32.09072
Paracou
m
0.0325
0.03250
0.03750
Code
read_tsv ("outputs/calib_str4.tsv" ) %>% mutate (sim = paste0 (site, "_" , a, "_" , b, "_" , m)) %>% left_join (best_par %>% slice_min (all_rmsep_norm)) %>% filter (! is.na (all_rmsep_norm)) %>% ggplot (aes (dbh_class, abundance)) + geom_line (aes (group = sim)) + geom_point (data = obs %>% filter (site == "Paracou" ), col = "red" ) + scale_y_log10 (labels = scales:: comma) + xlab ("DBH [ cm ]" ) + ylab (expression (Abundance~ "[" ~ ha^ {- ~ 1 }~ "]" )) + theme_bw () + facet_wrap (~ site)
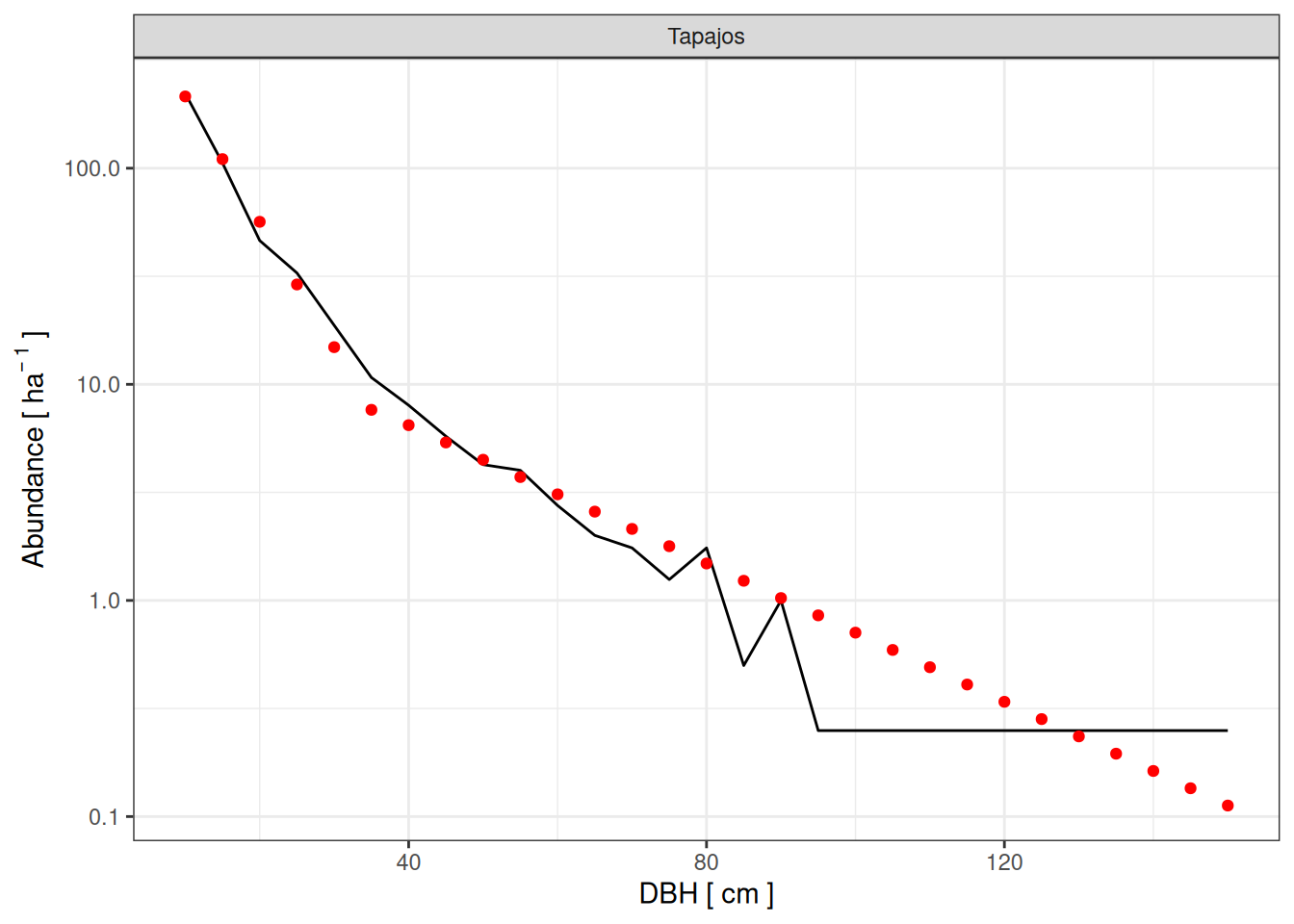
Tapajos
Code
<- read_tsv ("outputs/calib_str4.tsv" ) %>% filter (site == "Tapajos" ) %>% select (site, a, b, m) %>% unique () %>% mutate (dbh_class = list (seq (10 , 150 , by = 5 ))) %>% unnest (dbh_class) %>% left_join (read_tsv ("outputs/calib_str4.tsv" )) %>% select (- ba) %>% mutate_at (c ("abundance" , "agb" ), ~ ifelse (is.na (.), 0 , .)) %>% rename (simulated = abundance) %>% left_join (rename (obs, observed = abundance)) %>% mutate (observed = ifelse (is.na (observed), 0 , observed)) %>% group_by (site, a, b, m) %>% summarise (rmsep_dist = sqrt (mean ((observed - simulated)^ 2 )),rmsep_agb = sqrt ((143.7 - sum (agb)/ 10 ^ 3 / 2 )^ 2 ),rmsep_abund = sqrt ((470 - sum (simulated))^ 2 )%>% ungroup () %>% mutate (all_rmsep_norm = rmsep_dist/ 16.2 + rmsep_agb/ 143.7 + rmsep_abund/ 470 ) %>% mutate (sim = paste0 (site, "_" , a, "_" , b, "_" , m))<- eval_tap %>% group_by (site) %>% slice_min (all_rmsep_norm, n = 5 )
Code
read_tsv ("outputs/calib_str4.tsv" ) %>% mutate (sim = paste0 (site, "_" , a, "_" , b, "_" , m)) %>% left_join (best_tap) %>% filter (! is.na (all_rmsep_norm)) %>% group_by (site, a, b, m) %>% summarise (abundance = sum (abundance), agb = sum (agb)/ 10 ^ 3 / 2 ) %>% gather (variable, value, - site) %>% group_by (site, variable) %>% summarise (median = median (value), minimum = min (value), maximum = max (value)) %>% :: kable (caption = "Best structure parameters according to calibration with inventory data." )
Best structure parameters according to calibration with inventory data.
Tapajos
a
2.4500
2.3500
2.5000
Tapajos
abundance
467.0000
460.2500
484.2500
Tapajos
agb
144.8843
139.7424
148.6621
Tapajos
b
0.7565
0.6995
0.7850
Tapajos
m
0.0400
0.0300
0.0500
Code
read_tsv ("outputs/calib_str4.tsv" ) %>% mutate (sim = paste0 (site, "_" , a, "_" , b, "_" , m)) %>% left_join (best_tap %>% slice_min (all_rmsep_norm)) %>% filter (! is.na (all_rmsep_norm)) %>% ggplot (aes (dbh_class, abundance)) + geom_line (aes (group = sim)) + geom_point (data = obs %>% filter (site == "Tapajos" ), col = "red" ) + scale_y_log10 (labels = scales:: comma) + xlab ("DBH [ cm ]" ) + ylab (expression (Abundance~ "[" ~ ha^ {- ~ 1 }~ "]" )) + theme_bw () + facet_wrap (~ site)
All
Code
read_tsv ("outputs/calib_str4.tsv" ) %>% select (site, a, b, m) %>% unique () %>% mutate (dbh_class = list (seq (10 , 150 , by = 5 ))) %>% unnest (dbh_class) %>% left_join (read_tsv ("outputs/calib_str4.tsv" )) %>% select (- agb, - ba) %>% mutate (abundance = ifelse (is.na (abundance), 0 , abundance)) %>% rename (simulated = abundance) %>% left_join (rename (obs, observed = abundance)) %>% mutate (observed = ifelse (is.na (observed), 0 , observed)) %>% group_by (site, a, b, m) %>% summarise (normal = sqrt (mean ((observed - simulated)^ 2 )),lognormal = exp (sqrt (mean ((log (observed+ 1 ) - log (simulated+ 1 ))^ 2 )))) %>% gather (metric, rmsep, - site, - a, - b, - m) %>% group_by (site, metric) %>% slice_min (rmsep, n = 5 ) %>% mutate (rmsep = round (rmsep, 2 )) %>% summarise (RMSEP = paste0 (median (rmsep), " [" , min (rmsep), "-" , max (rmsep), "]" ),a = paste0 (median (a), " [" , min (a), "-" , max (a), "]" ),b = paste0 (median (b), " [" , min (b), "-" , max (b), "]" ),m = paste0 (median (m), " [" , min (m), "-" , max (m), "]" )%>% :: kable ()
Rows: 15261 Columns: 8
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (1): site
dbl (7): a, b, m, dbh_class, abundance, agb, ba
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Rows: 15261 Columns: 8
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (1): site
dbl (7): a, b, m, dbh_class, abundance, agb, ba
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Joining with `by = join_by(site, a, b, m, dbh_class)`
Joining with `by = join_by(site, dbh_class)`
`summarise()` has grouped output by 'site', 'a', 'b'. You can override using the `.groups` argument.
`summarise()` has grouped output by 'site'. You can override using the `.groups` argument.
Paracou
lognormal
1.12 [1.11-1.14]
1.8 [1.65-2]
0.386 [0.3005-0.55]
0.0325 [0.0325-0.035]
Paracou
normal
2.4 [1.38-2.7]
1.85 [1.8-1.9]
0.4145 [0.386-0.443]
0.0425 [0.0325-0.05]
Tapajos
lognormal
1.22 [1.19-1.22]
2.55 [2.5-2.7]
0.9135 [0.8635-0.9635]
0.0325 [0.03-0.045]
Tapajos
normal
2.54 [2.38-2.74]
2.35 [2.3-2.35]
0.6995 [0.671-0.6995]
0.045 [0.0375-0.05]
Code
bind_rows (%>% select (- sim) %>% gather (metric, rmsep, - site, - a, - b, - m) %>% mutate (metric = gsub ("rmsep_" , "" , metric)),%>% select (- sim) %>% gather (metric, rmsep, - site, - a, - b, - m) %>% mutate (metric = gsub ("rmsep_" , "" , metric))%>% group_by (site, metric) %>% slice_min (rmsep, n = 5 ) %>% mutate (rmsep = round (rmsep, 2 )) %>% summarise (RMSEP = paste0 (median (rmsep), " [" , min (rmsep), "-" , max (rmsep), "]" ),a = paste0 (median (a), " [" , min (a), "-" , max (a), "]" ),b = paste0 (median (b), " [" , min (b), "-" , max (b), "]" ),m = paste0 (median (m), " [" , min (m), "-" , max (m), "]" )%>% write_tsv ("figures/table_a4.tsv" )read_tsv ("figures/table_a4.tsv" ) %>% :: kable ()
Paracou
abund
5.75 [2-7.75]
1.75 [1.75-1.8]
0.3575 [0.3575-0.386]
0.0475 [0.0375-0.05]
Paracou
all_norm
0.16 [0.13-0.17]
1.8 [1.8-1.9]
0.386 [0.386-0.443]
0.0325 [0.0325-0.0375]
Paracou
ba
0.04 [0.03-0.07]
1.85 [1.65-2]
0.4715 [0.3505-0.5075]
0.0325 [0.03-0.05]
Paracou
dist
2.4 [1.38-2.7]
1.85 [1.8-1.9]
0.4145 [0.386-0.443]
0.0425 [0.0325-0.05]
Tapajos
abund
3 [0-3.5]
2.5 [2.4-2.65]
0.785 [0.728-0.9205]
0.035 [0.03-0.04]
Tapajos
agb
0.13 [0.04-0.19]
2.45 [2.35-2.5]
0.835 [0.6495-0.885]
0.045 [0.03-0.05]
Tapajos
all_norm
0.25 [0.18-0.25]
2.45 [2.35-2.5]
0.7565 [0.6995-0.785]
0.04 [0.03-0.05]
Tapajos
dist
2.54 [2.38-2.74]
2.35 [2.3-2.35]
0.6995 [0.671-0.6995]
0.045 [0.0375-0.05]
Code
bind_rows (%>% slice_min (all_rmsep_norm),%>% slice_min (all_rmsep_norm)%>% select (site, a, b, m) %>% mutate (berr = b + 0.39 - 0.57 * a) %>% kable ()
Paracou
1.80
0.3860
0.035
-0.25
Tapajos
2.45
0.7565
0.040
-0.25
Code
bind_rows (best_par, best_tap) %>% select (site, all_rmsep_norm, a, b, m) %>% gather (parameter, value, - site, - all_rmsep_norm) %>% group_by (site, parameter) %>% arrange (all_rmsep_norm) %>% summarise (minimum = min (value), median = median (value), best = first (value), maximum = max (value)) %>% kable ()
Paracou
a
1.8000
1.8000
1.8000
1.9000
Paracou
b
0.3860
0.3860
0.3860
0.4430
Paracou
m
0.0325
0.0325
0.0350
0.0375
Tapajos
a
2.3500
2.4500
2.4500
2.5000
Tapajos
b
0.6995
0.7565
0.7565
0.7850
Tapajos
m
0.0300
0.0400
0.0400
0.0500
Code
<- read_tsv ("outputs/calib_str4.tsv" ) %>% mutate (sim = paste0 (site, "_" , a, "_" , b, "_" , m)) %>% left_join (bind_rows (%>% slice_min (all_rmsep_norm),%>% slice_min (all_rmsep_norm)%>% filter (! is.na (all_rmsep_norm)) %>% ggplot (aes (dbh_class, abundance)) + geom_line (aes (group = sim, col = "TROLL" )) + geom_point (data = obs, aes (col = "field" )) + xlab ("DBH [ cm ]" ) + ylab (expression (Number~ of~ stems~ "[" ~ ha^ {- ~ 1 }~ "]" )) + theme_bw () + facet_wrap (~ site) + scale_colour_manual ("" , values = c ("#3f3f3f" , "#0da1f8" )) + theme (legend.position = "bottom" ) + geom_text (aes (col = "field" , label = lab), data = data.frame (dbh_class = 80 , abundance = 200 ,lab = c ("470 trees/ha & 287 Mg/ha" ,"612 trees/ha & 419 Mg/ha" ),site = c ("Tapajos" , "Paracou" ))) + geom_text (aes (col = "TROLL" , label = lab), data = data.frame (dbh_class = 80 , abundance = 180 ,lab = c ("471 trees/ha & 290 Mg/ha" ,"622 trees/ha & 413 Mg/ha" ),site = c ("Tapajos" , "Paracou" )))ggsave ("figures/f01.png" , g, dpi = 300 , width = 8 , height = 5 , bg = "white" )
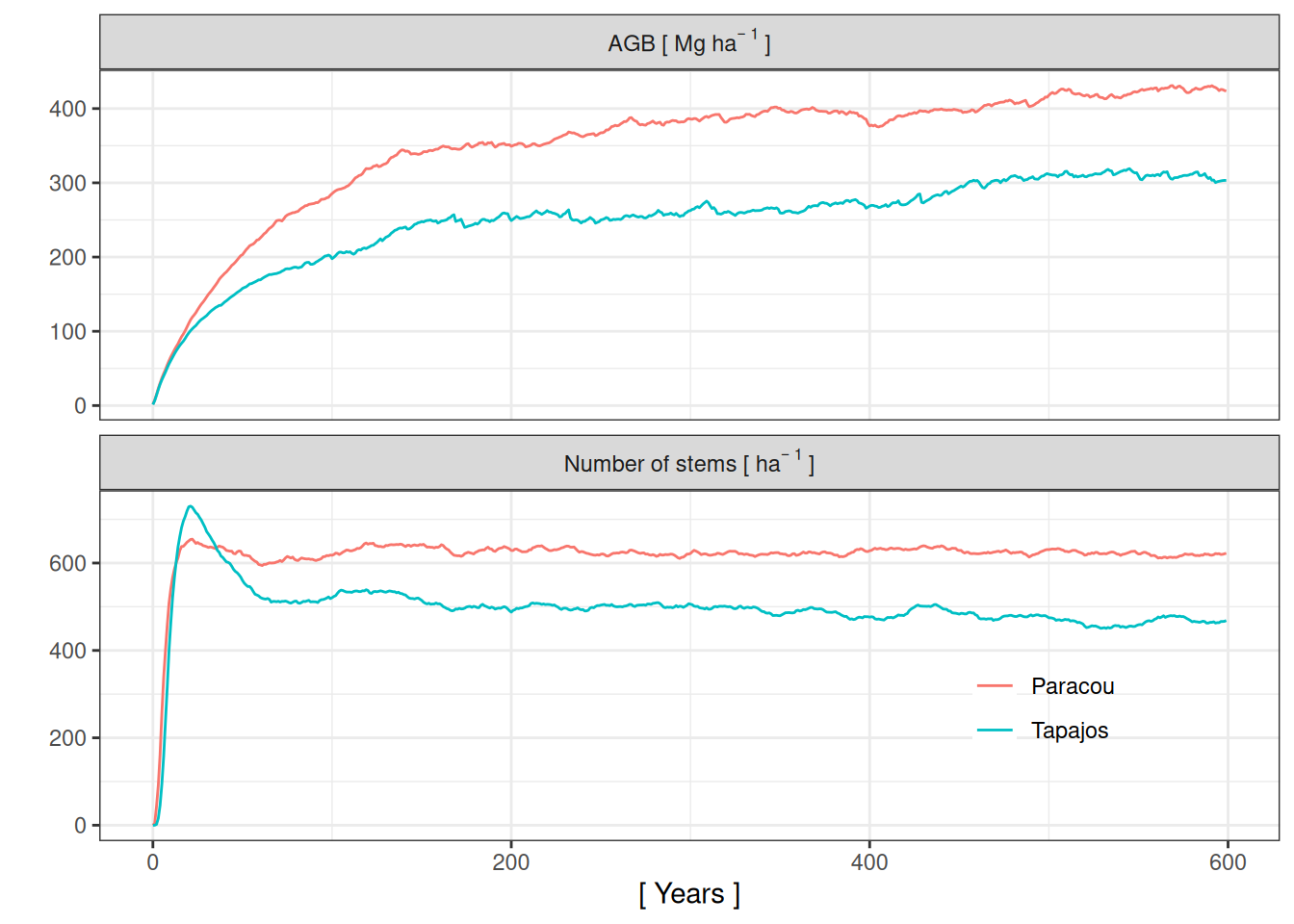
Spinup
Code
<- list (Paracou = read_tsv ("results/calib_str4_run/Paracou_1.8_-0.25_0.035/Paracou_1.8_-0.25_0.035_0_sumstats.txt" ) %>% mutate (date = as_date ("0000/01/01" )+ iter),Tapajos = read_tsv ("results/calib_str4_run/Tapajos_2.45_-0.25_0.04/Tapajos_2.45_-0.25_0.04_0_sumstats.txt" ) %>% mutate (date = as_date ("0000/01/01" )+ iter)%>% bind_rows (.id = "site" ) %>% group_by (site, year = year (date)) %>% summarise (abundance = mean (sum10), agb = mean (agb)/ 10 ^ 3 ) %>% gather (variable, value, - year, - site)<- spinup %>% mutate (var_long = recode (variable, "agb" = 'AGB~"["~Mg~ha^{-~1}~"]"' ,"abundance" = 'Number~of~stems~"["~ha^{-~1}~"]"' )) %>% ggplot (aes (year, value, col = site)) + geom_line () + facet_wrap (~ var_long, scales = "free_y" , nrow = 3 , labeller = label_parsed) + theme_bw () + xlab ("[ Years ]" ) + ylab ("" ) + scale_colour_discrete ("" ) + theme (legend.position = c (0.8 , 0.2 ), legend.background = element_blank ())ggsave ("figures/fa03.png" , g, dpi = 300 , width = 10 , height = 5 , bg = "white" )
Rice, Amy H., Elizabeth Hammond Pyle, Scott R. Saleska, Lucy Hutyra, Michael Palace, Michael Keller, Plínio B. de Camargo, Kleber Portilho, Dulcyana F. Marques, and Steven C. Wofsy. 2004.
“CARBON BALANCE AND VEGETATION DYNAMICS IN AN OLD- GROWTH AMAZONIAN FOREST.” Ecological Applications 14 (sp4): 55–71.
https://doi.org/10.1890/02-6006 .